Physical Address
304 North Cardinal St.
Dorchester Center, MA 02124
Enterobacter cloacae is a Gram-negative bacterium that plays a dual role as both a benign gut inhabitant and an opportunistic pathogen, often leading to serious infections in vulnerable populations.
Its identification is critical, particularly through biochemical tests like the indole test, which determines its ability to produce indole from tryptophan. While E. cloacae typically shows a negative result for this test, understanding its biochemical profile is essential for effective clinical management.
As we explore this fascinating bacterium, the intricate relationship between humans and microbes unfolds, revealing challenges and insights that shape modern medicine.
Contents
Enterobacter is a genus of bacteria that belongs to the family Enterobacteriaceae, which includes a diverse group of gram-negative, rod-shaped organisms. These bacteria are commonly found in the intestines of humans and animals, as well as in various environmental sources such as soil and water.
Enterobacteriaceae Scientific Name: The scientific name for the family commonly known as Enterobacteriaceae is Enterobacteriaceae Rahn, 1937. This family is classified under the domain Bacteria, phylum Pseudomonadota, class Gammaproteobacteria, and order Enterobacterales.
Enterobacteriaceae includes over 30 genera and more than 100 species, many of which are significant both as commensals in the intestines of animals and as pathogens, such as Escherichia coli, Salmonella, Klebsiella, and Shigella.
The etymology of the name comes from “enterobacterium,” indicating its association with the intestines (from Greek “enteron”) and the suffix “-aceae” designating it as a family
Scientific Classification:
Pathogenic Potential:
Environmental Presence:
While many species are harmless or beneficial, some can act as opportunistic pathogens, particularly in immunocompromised individuals. Enterobacter’s classification and characteristics is essential for both clinical and environmental microbiology.
Enterobacter cloacae is a significant bacterium within the Enterobacteriaceae family, known for its rod-shaped morphology and gram-negative characteristics. This organism is commonly found in various environments, including human intestines, soil, and water.
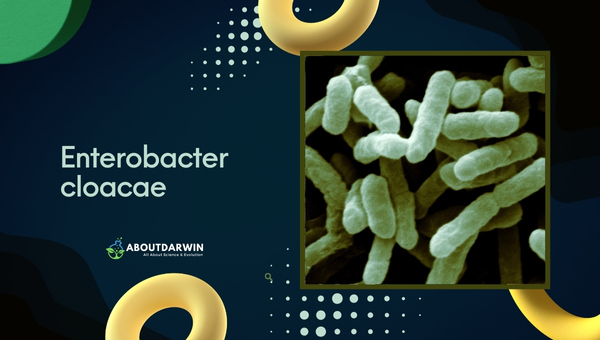
It plays a dual role as both a member of the normal gut flora and an opportunistic pathogen, particularly in healthcare settings. Its properties and characteristics is crucial for managing infections it may cause, especially given its increasing antibiotic resistance.
| Characteristic | Property |
|---|---|
| Capsule | Negative (-ve) |
| Catalase | Positive (+ve) |
| Citrate Utilization | Positive (+ve) |
| Flagella | Positive (+ve) |
| Gas from Glucose | Positive (+ve) |
| Gelatin Hydrolysis | Negative (-ve) |
| Gram Staining | Negative (-ve) |
| Growth in KCN | Positive (+ve) |
| H2S Production | Negative (-ve) |
| Indole Production | Negative (-ve) |
| Motility | Motile |
| Methyl Red (MR) | Negative (-ve) |
| Nitrate Reduction | Positive (+ve) |
| Oxidase Test | Negative (-ve) |
| Oxidative-Fermentative (OF) | Facultative Anaerobes |
Morphology and Biochemical Properties:
Enterobacter cloacae is a rod-shaped, gram-negative bacterium measuring approximately 0.3-0.6 x 0.8-2.0 μm. It is facultatively anaerobic, meaning it can grow in both the presence and absence of oxygen, utilizing aerobic respiration or fermentation for ATP production.
The bacterium is oxidase-negative and catalase-positive, indicating its metabolic capabilities. It possesses peritrichous flagella, which facilitate motility and adhesion to surfaces.
Pathogenicity:
E. cloacae is recognized as a nosocomial pathogen associated with various infections, including bacteremia, urinary tract infections (UTIs), lower respiratory tract infections, and intra-abdominal infections.
It poses a higher risk to vulnerable populations such as the elderly and immunocompromised individuals. The organism’s ability to form biofilms contributes to its pathogenicity and resistance to host defenses.
Antibiotic Resistance:
This bacterium exhibits significant antibiotic resistance due to the production of beta-lactamases, particularly AmpC beta-lactamases, which confer resistance to many commonly used antibiotics, including third-generation cephalosporins.
The genetic mechanisms behind this resistance complicate treatment options and pose challenges in clinical settings. E. cloacae’s intrinsic resistance to ampicillin and other beta-lactam antibiotics further complicates management strategies.
Environmental Role:
Apart from its clinical significance, E. cloacae plays a role in environmental bioremediation. It has been utilized in biodegradation processes for pollutants such as explosives and plastics like polyvinyl alcohol (PVA), showcasing its versatility beyond pathogenicity.
Clinical Relevance:
The increasing prevalence of antibiotic-resistant strains of E. cloacae has led to its classification as a priority pathogen by health organizations aiming to develop new antibiotics.
Its presence in clinical specimens underscores the need for ongoing surveillance and research into effective treatment protocols.
Also Read: Proteus Mirabilis: Understanding Biochemical Identification
Enterobacter cloacae is a rod-shaped, gram-negative bacterium belonging to the Enterobacteriaceae family. This organism is oxidase-negative and catalase-positive, indicating it does not produce cytochrome c oxidase but does produce catalase.
Its resistance to multiple antibiotics, including beta-lactam antibiotics due to the production of beta-lactamases, complicates treatment options. The oxidase test is crucial for differentiating E. cloacae from other Enterobacter species in microbiological diagnostics.
Enterobacter cloacae is a significant bacterium within the Enterobacteriaceae family, known for its role in various infections, particularly in immunocompromised individuals.
Understanding its biochemical characteristics is crucial for identification and treatment in clinical settings. The following outlines the key biochemical tests used to identify Enterobacter cloacae, providing essential insights into its metabolic capabilities and resistance patterns.
These biochemical tests are critical for the identification and management of Enterobacter cloacae infections in clinical microbiology.
The Enterobacter cloacae complex (ECC) comprises several closely related species, including Enterobacter cloacae, which are significant in clinical settings due to their role as opportunistic pathogens.
These bacteria are known for their multidrug resistance, particularly to beta-lactam antibiotics, making treatment challenging. The ability of E. cloacae to ferment lactose is a key characteristic that aids in its identification in laboratory settings.
The treatment options and resistance mechanisms is crucial for managing infections caused by this complex.
Treatment of infections caused by Enterobacter cloacae typically involves the use of beta-lactam antibiotics, such as cefepime and gentamicin, especially in cases where the strain is not resistant.
However, due to the high prevalence of multidrug-resistant strains within the E. cloacae complex, susceptibility testing is essential before initiating therapy to ensure effective treatment options are chosen.
The Enterobacter cloacae complex includes several species, with E. cloacae and E. hormaechei being the most frequently isolated in clinical specimens.
These bacteria can cause a variety of infections, including urinary tract infections and septicemia, particularly in immunocompromised patients.
Their ability to produce AmpC β-lactamases contributes to their resistance against many antibiotics, complicating treatment efforts.
A notable feature of the Enterobacter cloacae complex is its ability to ferment lactose, which is often used as a diagnostic trait in microbiological laboratories.
This fermentation capability can help differentiate E. cloacae from other non-lactose fermenting enteric bacteria, aiding in accurate identification and subsequent treatment decisions.
Also Read: Arteries and Veins: Understanding Differences and Impacts on Health
The field of microbiology is continuously evolving, leading to innovative methods for identifying pathogens such as Enterobacter cloacae.
This bacterium, a member of the Enterobacteriaceae family, can be challenging to identify using traditional biochemical tests due to its complex taxonomy.
Genomic Sequencing has revolutionized our understanding of bacterial identification. This technique allows researchers to analyze the complete genetic material of E. cloacae, revealing unique sequences that serve as genetic fingerprints for different species.
Whole Genome Sequencing (WGS) is particularly effective in distinguishing closely related bacterial species, a task that conventional biochemical tests often struggle with.
By employing genomic sequencing, scientists can gain insights into the evolutionary adaptations of E. cloacae, including its potential vulnerabilities to treatments.
This knowledge is crucial for developing targeted therapies and understanding how this bacterium may evolve resistance to antibiotics.
Matrix-Assisted Laser Desorption/Ionization Time-of-Flight (MALDI-TOF) Mass Spectrometry is another cutting-edge technique that has gained traction in clinical microbiology.
This method utilizes laser technology to ionize bacterial proteins, which are then analyzed based on their unique spectral signatures. Each bacterium produces a distinct pattern when subjected to this analysis, allowing for precise identification through sophisticated software.
MALDI-TOF MS offers several advantages over traditional biochemical tests, including significantly faster results without sacrificing accuracy.
Recent studies have shown that when combined with an extensive online database and advanced algorithms, MALDI-TOF MS can achieve an identification rate of up to 92% for E. cloacae complex isolates.
This efficiency is critical in clinical settings where timely identification can impact patient management and treatment outcomes.
The advancements in identification techniques such as genomic sequencing and MALDI-TOF mass spectrometry exemplify the significant progress made in microbiology.
These methods not only improve the accuracy of identifying Enterobacter cloacae but also enhance our understanding of its biology and resistance mechanisms.
As these technologies continue to develop, they will play an essential role in combating infections caused by this opportunistic pathogen, underscoring the importance of accurate microbial identification in healthcare settings.
These are the Importance of Identifying Enterobacter Cloacae :
Overall, identifying Enterobacter cloacae accurately shapes both individual patient care and broader healthcare landscape actions.
Also Read: Streptococcus Pneumoniae: Identification & Biochemical Tests
Enterobacter usually has a negative methyl-red test, a positive Voges-Proskauer test, can use citrate as a carbon source, can grow in Moller’s KCN medium at 30 °C, and are ornithine positive, although ‘Enterobacter agglomerans complex’ is ornithine decarboxylase negative.
The gold standard for diagnosing Enterobacter infections is the utilization of cultures. It is recommended that at least two sets of blood cultures be obtained, one aerobic and one anaerobic bottle. MacConkey agar can be used to determine if the specimen is lactose fermenting.
The genus Enterobacter ferments lactose with gas production during a 48-hour incubation at 35-37 °C in the presence of bile salts and detergents. It is oxidase-negative, indole-negative, and urease-variable.
The most important test to document Enterobacter infections is culture. Direct Gram staining of the specimen is also very useful because it allows rapid diagnosis of an infection caused by gram-negative bacilli and helps in the selection of antibiotics with known activity against most of these bacteria.
The treatment of Enterobacter cloacae infections requires precise identification and tailored therapeutic strategies. This bacterium is known for its resistance to multiple antibiotics, making effective treatment challenging.
Clinicians often rely on biochemical tests and advanced techniques like genomic sequencing and MALDI-TOF mass spectrometry to accurately diagnose infections.
The specific strain and its resistance patterns is crucial for selecting appropriate antibiotics, which can significantly improve patient outcomes. Ultimately, ongoing research and innovation in diagnostic methods will enhance our ability to combat this opportunistic pathogen effectively.